Drawing Inference
Lucy D’Agostino McGowan
Data
When is a simple linear regression model a useful descriptive summary?
- Linearity holds
- The residuals have “zero mean” (this is always true!)
- The datapoints are independent
What if we want to draw inference on another sample?
Inference
- So far we’ve only been able to describe our sample
- For example, we’ve just been describing \(\hat{\beta}_1\) the estimated slope of the relationship between \(x\) and \(y\)
- What if we want to extend these claims to the population?
Magnolia data
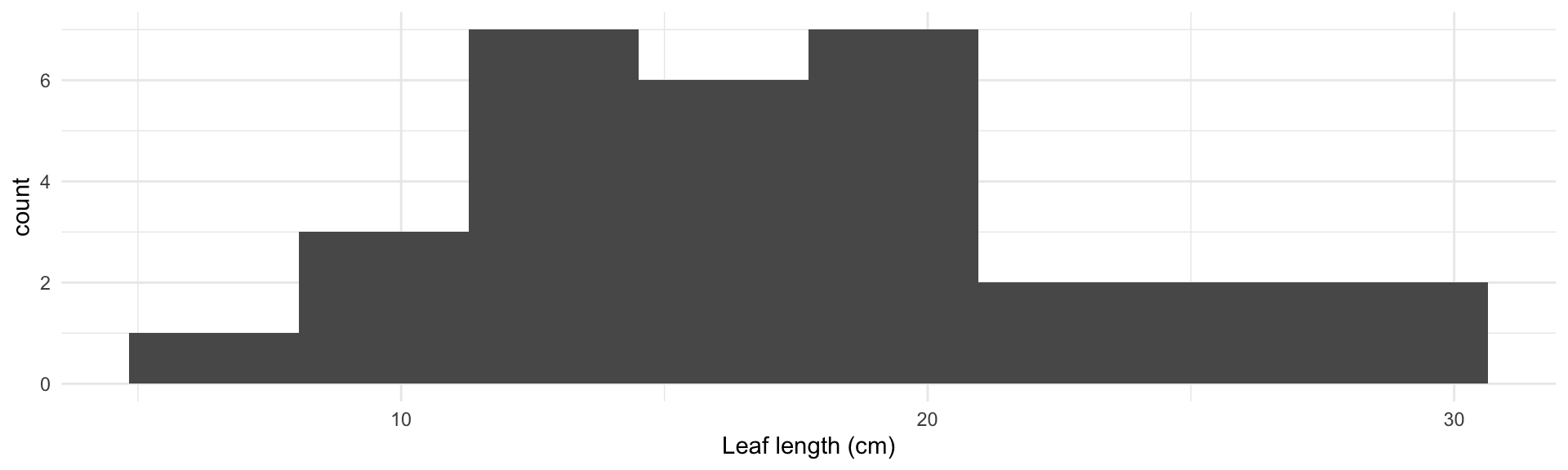
How can I visualize a single continuous variable?
Magnolia data
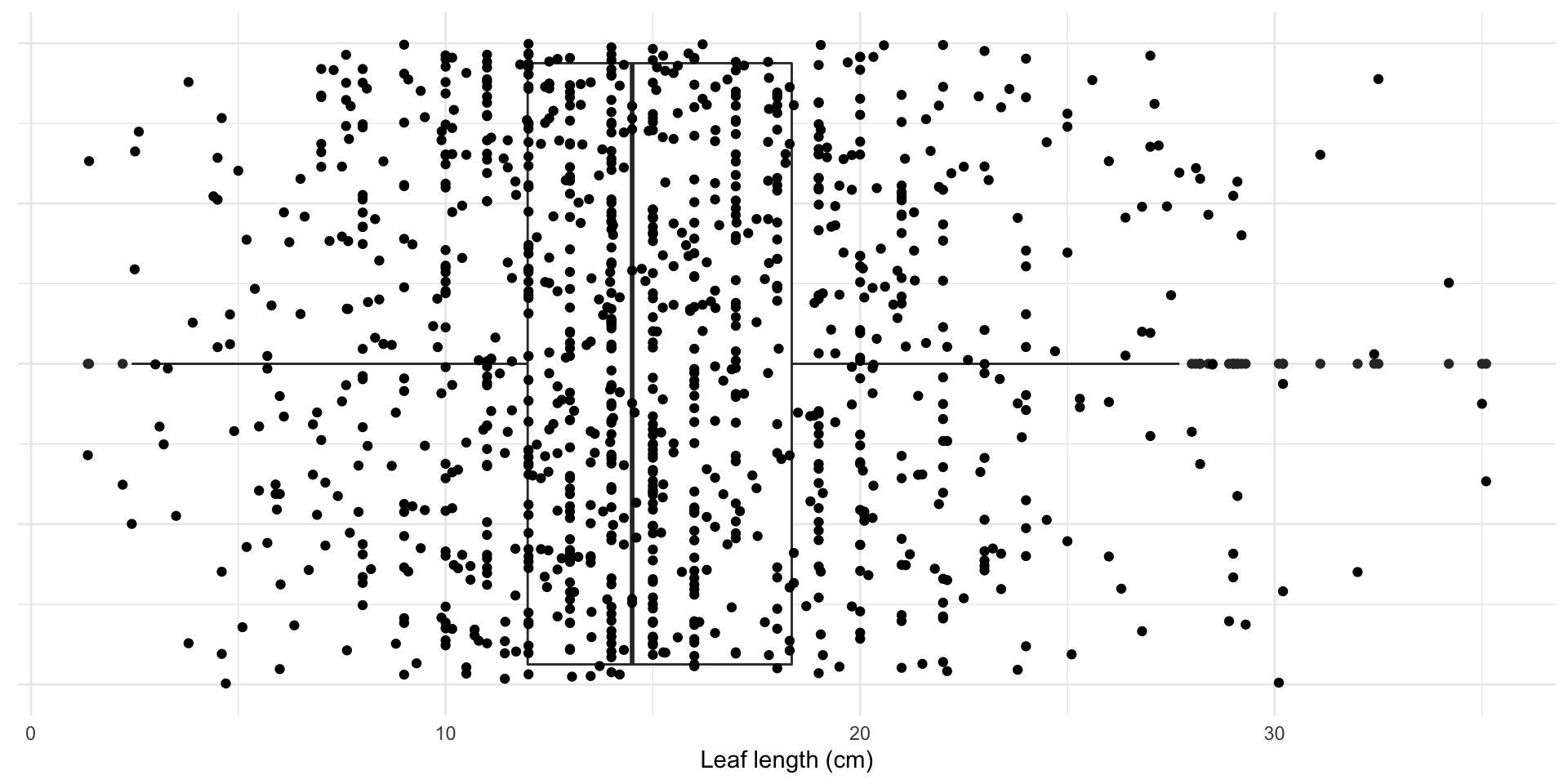
Magnolia data
Code
set.seed(1)
ggplot(full_magnolia_data,
aes(x = leaf_length, y = 1)) +
geom_boxplot() +
geom_jitter() +
geom_jitter(data = magnolia_data, color = "cornflower blue", size = 3) +
labs(x = "Leaf length (cm)") +
theme(axis.title.y = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank())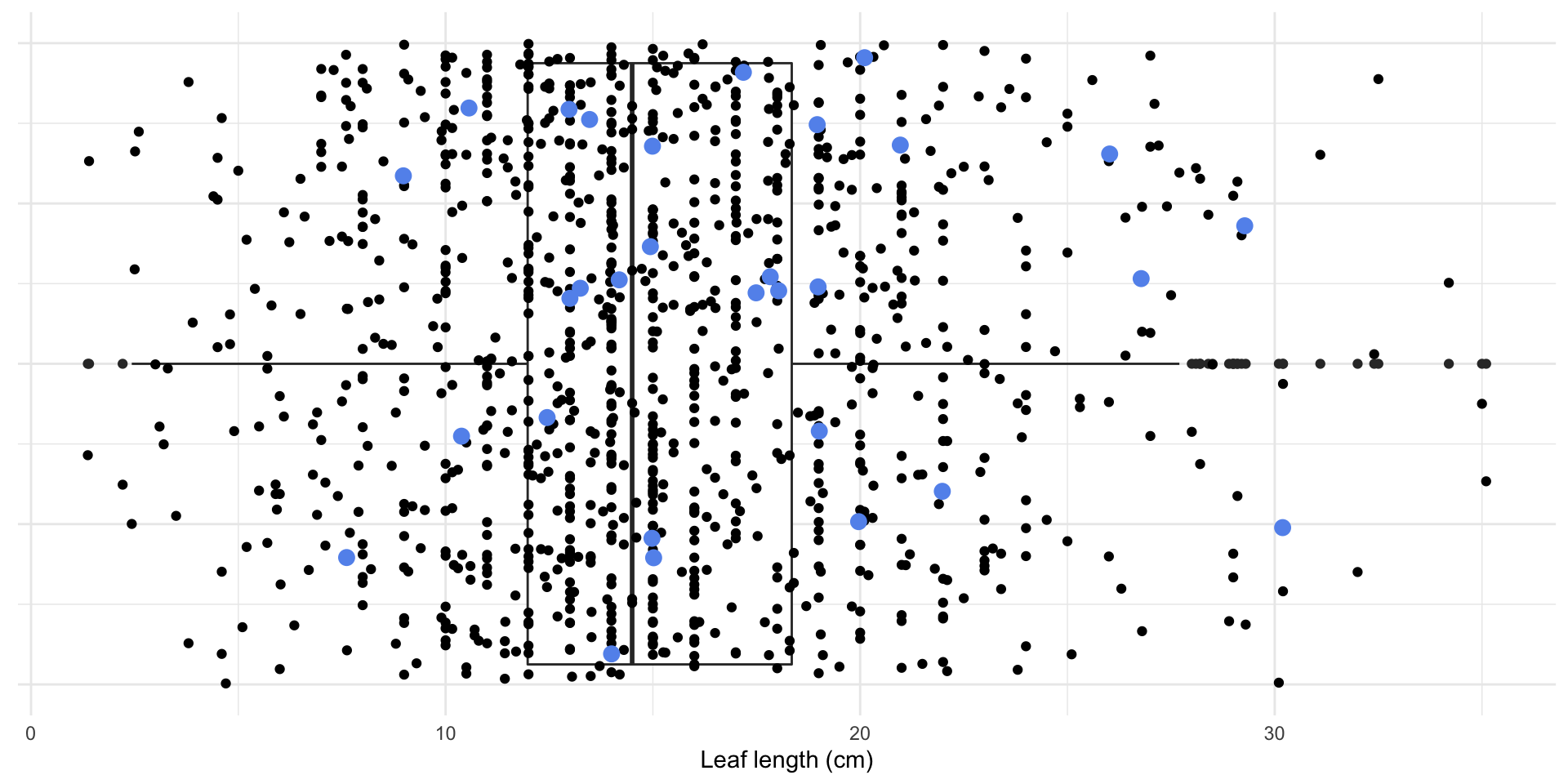
Magnolia data
Magnolia data
How can I calculate the average leaf length of the magnolias in my sample in R?
Magnolia data
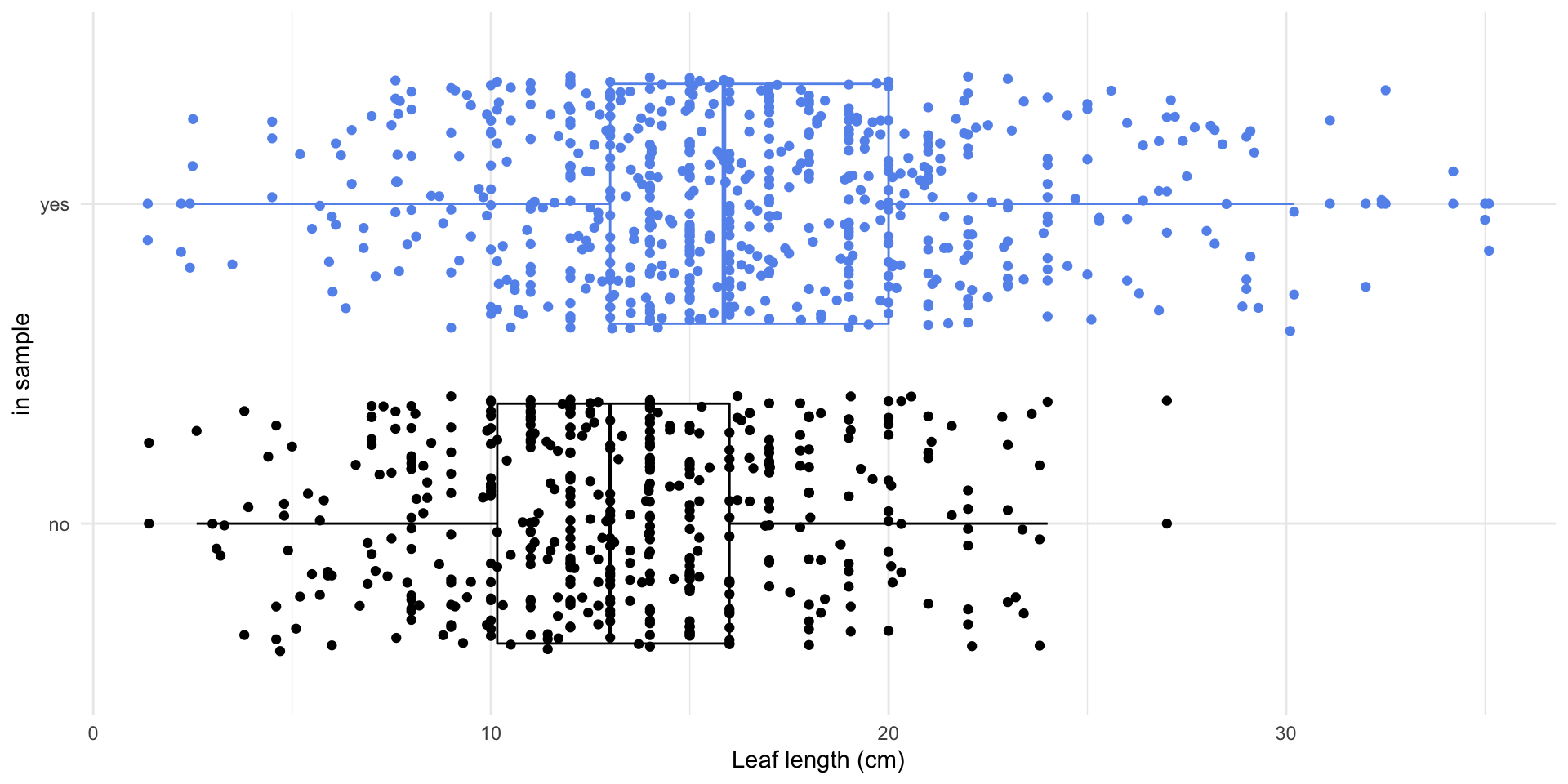
What if I want to know the average leaf length of the magnolias on the Mag Quad?
How can we quantify how much we’d expect the mean to differ from one random sample to another?
- We need a measure of uncertainty
- How about the standard error of the mean?
- The standard error is how much we expect the sample mean to vary from one random sample to another.
Magnolia data
How can we quantify how much we’d expect the mean to differ from one random sample to another?
Call:
lm(formula = leaf_length ~ 1, data = magnolia_data)
Residuals:
Min 1Q Median 3Q Max
-9.509 -3.809 -1.029 2.621 13.071
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 17.129 1.031 16.62 2.32e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 5.645 on 29 degrees of freedom Application Exercise
- Create a new project from this template in RStudio Pro:
- Replace the text in the top code chunk that says
INSERT YOUR GOOGLE SPREADSHEET URL HEREwith the Google Spreadsheet URL with your magnolia data. - Fit an intercept only model to calculate the average leaf length in your sample
- Use the
summaryfunction on the linear model you fit - What is the standard error for the mean length? Interpret this value.
05:00
confidence intervals
If we use the same sampling method to select different samples and computed an interval estimate for each sample, we would expect the true population parameter (the average leaf length on the Mag Quad) to fall within the interval estimates 95% of the time.
Confidence interval
\[\bar{x} \pm t^∗ \times SE_{\bar{x}}\]
- \(t^*\) is the critical value for the \(t_{n−1}\) density curve to obtain the desired confidence level
- Often we want a 95% confidence level.
Demo
Let’s do it in R!
Call:
lm(formula = leaf_length ~ 1, data = magnolia_data)
Residuals:
Min 1Q Median 3Q Max
-9.509 -3.809 -1.029 2.621 13.071
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 17.129 1.031 16.62 2.32e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 5.645 on 29 degrees of freedomLet’s do it in R!
Why 0.025?
Call:
lm(formula = leaf_length ~ 1, data = magnolia_data)
Residuals:
Min 1Q Median 3Q Max
-9.509 -3.809 -1.029 2.621 13.071
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 17.129 1.031 16.62 2.32e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 5.645 on 29 degrees of freedomLet’s do it in R!
Why lower.tail = FALSE?
Call:
lm(formula = leaf_length ~ 1, data = magnolia_data)
Residuals:
Min 1Q Median 3Q Max
-9.509 -3.809 -1.029 2.621 13.071
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 17.129 1.031 16.62 2.32e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 5.645 on 29 degrees of freedomLet’s do it in R!
Call:
lm(formula = leaf_length ~ 1, data = magnolia_data)
Residuals:
Min 1Q Median 3Q Max
-9.509 -3.809 -1.029 2.621 13.071
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 17.129 1.031 16.62 2.32e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 5.645 on 29 degrees of freedom[1] 16.7[1] 13confidence intervals
If we use the same sampling method to select different samples and computed an interval estimate for each sample, we would expect the true population parameter (the mean) to fall within the interval estimates 95% of the time.
Application Exercise
- Open
appex-08.qmd - Calculate the \(t^*\) value for your confidence interval
- Calculate the confidence interval “by hand” using the \(t^*\) value from exercise 2 and the mean and standard error from the previous application exercise
- Calculate the confidence interval using the
confintfunction - Interpret this value
05:00